Vendor centroiding is not supported for Waters ion mobility data due to the complex, high-resolution profile format used by Waters instruments. This prevents tools like MSConvert from applying centroiding during data conversion. Researchers should use profile-mode workflows or third-party software to analyze Waters IMS datasets effectively.
Stay tuned with us — we will talk in detail about vendor centroiding is not supported for Waters ion mobility data, its causes, workarounds, and the best tools for successful data analysis.
What Does “Vendor Centroiding Is Not Supported for Waters Ion Mobility Data” Mean?
If you’ve ever tried to process Waters ion mobility spectrometry (IMS) data and encountered the message “vendor centroiding is not supported for waters ion mobility data,” you’re not alone. This statement refers to a data processing limitation where software such as ProteoWizard’s MSConvert cannot apply vendor-specific centroiding algorithms to Waters IMS datasets.
In mass spectrometry, centroiding is a process that reduces LC-MS raw data size and complexity by summarizing each detected ion peak into a single point, rather than preserving the complete profile. Many vendors provide proprietary centroiding algorithms embedded in their hardware or software systems. However, in the case of Waters, such support is not extended to ion mobility data.
This limitation creates hurdles in many standard workflows, especially for users trying to Convert Waters raw to mzML for downstream analysis tools that require centroided data. For researchers dealing with large-scale proteomics or metabolomics projects, understanding what this error means and how to work around it is crucial.
In essence, the error message is a signal that your chosen software tool cannot process the data in its current form using the vendor’s optimized algorithms. Instead, alternative processing steps are required.
Why Waters Ion Mobility Data Cannot Be Vendor-Centric
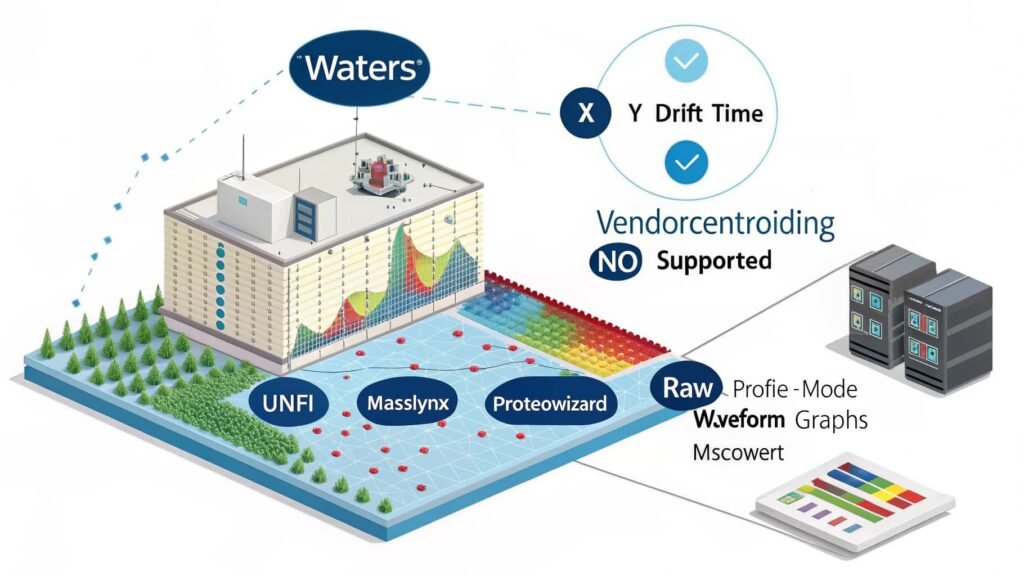
The architecture of Waters raw file format is one of the primary reasons why vendor centroiding is not supported. Unlike traditional mass spectrometry data, ion mobility data adds a third dimension to the dataset, representing drift time. This results in a far more complex data matrix, which makes it difficult to apply traditional centroiding approaches.
Waters typically stores its data in profile mode to retain the highest possible resolution. The vendor believes that this method provides a more accurate and detailed representation of each ion’s behavior, especially when accounting for mobility separation. Therefore, no built-in centroiding algorithms are applied to ion mobility dimensions within their raw data files.
Furthermore, Waters utilizes proprietary data formats such as .raw, which are not fully transparent or easily manipulated outside of their software ecosystem. These files do not allow third-party applications like ProteoWizard to access or execute vendor-specific centroiding methods.
Even when working with Waters’ official software like UNIFI or MassLynx, users often find that centroiding is limited or not possible in specific workflows. The company prioritizes full-resolution analysis, which benefits specific high-precision applications but limits interoperability.
Tools Affected by the Centroiding Limitation
The limitation affects a range of data processing and conversion tools commonly used in proteomics and mass spectrometry. Chief among these is ProteoWizard’s MSConvert, one of the most popular open-source tools for converting mass spectrometry data into open formats like mzML or mzXML.
When users attempt to convert Waters IMS data using MSConvert, they often enable the centroiding option for reducing file size and optimizing downstream processing. However, due to the lack of vendor support, the software returns the error message in question and skips centroiding entirely for IMS dimensions.
Other affected tools include:
- MaxQuant: Often expects centroided data for faster processing.
- Skyline: May show limited support for profile-mode-only data. Some users report needing to select “chambm pwiz skyline I agree to the vendor licenses” during setup to access proprietary conversion features.
- OpenMS: Can be used for centroiding but may introduce errors without proper configuration.
The absence of vendor centroiding impacts pipeline efficiency and may necessitate significant changes to the user’s data preprocessing strategy. Additionally, storage requirements increase, as profile-mode data consumes considerably more disk space.
How to Work with Waters IMS Data Without Vendor Centroiding
Despite this limitation, there are several effective strategies for working with Waters ion mobility data. The most straightforward approach is to accept and process the data in its original profile mode.
Many modern tools now support profile-mode analysis, and in some cases, retaining the full resolution of the raw data leads to better identification and quantification. For instance, DIA-NN and Spectronaut can handle Waters IMS data without requiring centroiding, offering advanced algorithms that are optimized for profile-mode input.
Another approach is to fine-tune your use of MSConvert. By disabling the centroiding option and ensuring the output remains in profile mode, you can Convert Waters raw to mzML files that retain compatibility with tools that accept profile data.
Additionally, users can explore developing custom workflows using Python or R, leveraging libraries like pyOpenMS or mzR to work with high-resolution profile data. These tools may require more setup but offer granular control over data handling and interpretation.
Workarounds and Alternative Centroiding Methods
If centroiding is necessary for your workflow, third-party software-based centroiding methods offer a possible solution. However, these approaches should be used cautiously, as they may not match the accuracy of vendor-optimized algorithms.
Mzmine Ion Mobility Support
Mzmine ion mobility features have improved in recent versions, making it a viable tool for handling Waters IMS data. Users can apply peak detection and deconvolution on profile-mode files within MZmine. Recent developments in Reproducible mass spectrometry data processing and compound annotation in MZmine 3 have demonstrated the platform’s strength in dealing with complex mobility-separated data.
Users can also employ Python-based centroiding through packages like ms_deisotope or pyteomics, though this again requires a strong understanding of data structures and algorithm parameters.
If you’re dealing with different formats, you can also use tools to Convert mgf to mzML, ensuring compatibility with analysis platforms that require mzML.
It’s essential to validate these results carefully, mainly when the data is used for quantitative studies. Artifacts introduced during software centroiding can skew peak intensities or misrepresent the ion mobility separation.
Ultimately, the best workaround often involves avoiding centroiding altogether and redesigning the pipeline to accommodate profile-mode data.
Real-World Examples and Researcher Feedback
Many researchers have shared their experiences with this issue on platforms like GitHub, StackExchange, and vendor-specific forums. In the proteomics community, it’s widely known that “vendor centroiding is not supported for waters ion mobility data” is a recurring roadblock.
For example, a GitHub user working with ProteoWizard reported consistent failures when attempting to centroid Waters Synapt G2-S data. After extensive testing, the user confirmed that processing in profile mode not only resolved the issue but also improved identification rates when analyzed with DIA-NN.
Similarly, university labs that rely heavily on Skyline have noted that adapting to profile-mode workflows was challenging but ultimately worthwhile. One researcher from a UK-based proteomics core facility stated that they transitioned their entire Waters data processing to profile-mode pipelines with minimal long-term drawbacks.
Case studies have also shown that tools like Spectronaut can handle the full complexity of Waters IMS data without requiring centroiding, producing highly reproducible results.
These community insights help validate the technical recommendations and provide practical reassurance that viable alternatives exist.
Official Recommendations from Waters and Software Developers
Waters has acknowledged this limitation in their documentation and support channels. The official stance is that ion mobility data is best used in profile mode to preserve its rich structure and detail. Consequently, the company does not provide vendor centroiding for this data type.
In technical documentation for tools like MassLynx and UNIFI, Waters suggests workflows that emphasize full-resolution data processing. They encourage users to rely on proprietary software for visualization and simple quantification but recommend external tools for more advanced analyses.
On the developer side, teams behind ProteoWizard, Skyline, and OpenMS have included disclaimers and configuration notes in their documentation. ProteoWizard, for example, logs a warning message indicating that centroiding is not possible for Waters IMS data and proceeds to process the file in profile mode.
These clear positions from both the vendor and software developers reinforce the idea that researchers should shift expectations and adapt workflows rather than seek to bypass the limitation.
Will Vendor Centricity Ever Be Supported in the Future?

As of now, there is no public roadmap indicating that Waters plans to introduce vendor centroiding for ion mobility data. The company’s commitment to high-fidelity data storage means they are unlikely to change their stance unless there is a significant shift in user demand or industry standards.
However, pressure from the scientific community and increasing adoption of open formats like mzML with IM dimension support may push vendors toward more flexible solutions. Collaboration between tool developers and vendors could lead to standardized centroiding frameworks that work across multiple platforms.
In the meantime, researchers must continue relying on profile-mode workflows or third-party centroiding algorithms. It remains critical for tool developers to enhance support for Waters’ raw file format and provide better documentation to help users navigate these limitations.
While nothing is guaranteed, the continued evolution of open-source mass spectrometry tools could eventually lead to more robust centroiding solutions for ion mobility data, even without direct vendor involvement.
Final Thoughts on Handling Waters Ion Mobility Data Without Vendor Centroiding
The message “vendor centroiding is not supported for waters ion mobility data” can be frustrating, but it’s not a dead end. With modern tools capable of processing full-resolution profile data, researchers have options to maintain data quality and integrity.
By understanding the reasons behind the limitations and adapting workflows accordingly, it’s possible to extract meaningful insights from complex Waters datasets. Whether by using DIA-NN, Spectronaut, Mzmine ion mobility, or even writing custom scripts, success lies in embracing the profile-mode paradigm.
Ultimately, this limitation is a reminder of the importance of flexible, open scientific tools that can accommodate the full range of vendor data types without compromising performance.
Frequently Asked Questions (FAQ)
1. Can I force vendor centroiding on Waters IMS data using MSConvert?
No, MSConvert cannot perform vendor centroiding on Waters ion mobility data. It will log an error and skip centroiding for IMS dimensions.
2. What is the best alternative to vendor centroiding for Waters data?
Using profile-mode workflows in tools like DIA-NN, Spectronaut, or Mzmine ion mobility is currently the best approach. These tools handle Waters IMS data without requiring centroiding.
3. Is it safe to apply third-party centroiding algorithms to Waters data?
You can apply third-party centroiding using tools like OpenMS or Mzmine, but you must validate results carefully to avoid data artifacts.
4. Why does Waters store ion mobility data in profile mode only?
Waters prioritizes data resolution and accuracy, believing profile mode best represents ion mobility behavior. As a result, they do not support centroiding natively.
5. Are there any plans from Waters to add centroiding support for IMS data?
As of now, Waters has not announced any plans to support vendor centroiding for ion mobility data.
Also read:
- Widner Mobility Oscillator Guide for Engineers & Traders!
- xfinity mobile customer service phone number get human!
- How to Delete FB Account Permanently from Mobile!
